vtkbioeng is a C++ class library built on the Visualisation Toolkit (VTK). It contains classes that were needed for our research that were not catered for in the main VTK library. The most popular classes are the collision detection filter and cell removal filter. The library is hosted on sourceforge.net or on github
Here’s a description of a couple of the open sourced classes:
vtkCollisionDetectionFilter
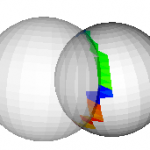
This filter takes two polydata inputs and performs a collision detection routine, using oriented bounding boxes (vtkOBBTree). The input polydata are passed to the output ports 0 and 1, with the intersecting triangles colored if the “GenerateScalars” flag is set. The intersecting triangles are colored in the order they are found… red first, blue last (left figure)
A list of the contacting cells can be obtained as a vtkIdTypeArray using “GetContactCells(int i)”
The output from port 2 is a polydata containing the points of intersection. “GetContactsOutputPort()” is a convenience method for “GetOutputPort(2)”
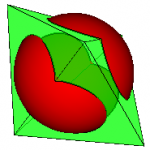
This “line of intersection” can be used to clip one surface with another. The centre figure shows a sphere being clipped by a bipyramid
vtkPolylineCurvature
This class is not in the current vtkBioeng package – there are a couple more curvature classes that will all be added together.
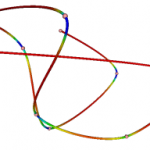
Uses Frenet frames to calculate the curvature of a polyline. The torsion doesn’t seem to be correct in testing. vtkSplineFilter can be use to re-parameterize the polyline before using this class.
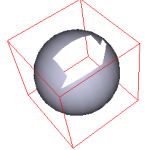
vtkRemoveCellsFilter
Along with vtkCellPicker, cells on a polydata surface can be interactively picked and deleted. As a cell is deleted the cell ids are remapped to compensate for the deleted cell… this classes keeps track of the remapping so that the correct cells are subsequently deleted. This class is now a subclass of vtkCellsFilter. It also has a sister class vtkSelectCellsFilter which deletes the cells unmarked by the user. You can think of it as the inverse of vtkRemoveCellsFilter.